The problem
I am in the process of migrating my (rather ugly) small blog from “Bloggers” to `blogdown` and, as several others, I choose to use the hugo-academic theme due to its good looks, simplicity, and “focus” towards researchers.
One nice feature of
hugo-academic is that it includes out-of-the-box a “Publications” section, allowing researchers to easily create a list of their publication as a section of the website.
Unfortunately, in order to populate that list, users have to manually create one different
.md file for each publication, by cutting and pasting several different info (e.g., title, authors, etc.) in a "simple" template like this one.
Since I was not in the mood of doing that, and no automatic solutions could be found (well, there appears to be a
python one, but we are speaking R, here…), I decided to try and develop some script to automatically create the required md files starting from a BibTex list of my publications. Here are the results of that effort.A possible solution
Preparing the BibTex file
To automatically create the publications
md files, all you need is a (properly formatted) BibTex file. Since I did not have one ready, I created mine by exporting my publications list from Scopus, but you could use any valid BibTeX file.
One important thing, though, is that you have to be sure that the file is saved with UTF-8 encoding. If you are not sure, you can open it in RStudio (or any decent text editor), and then re-save it with a forced encoding (in RStudio, you can use
File-->Save with Encoding)Creating an import script
Now, you need a script that reads the
BibTex entries and use the data to populate one different mdfile for each publication. Below you can find my attempt at this.bibtex_2academic <- function(bibfile,
outfold,
abstract = FALSE,
overwrite = FALSE) {
require(RefManageR)
require(dplyr)
require(stringr)
require(anytime)
# Import the bibtex file and convert to data.frame
mypubs <- ReadBib(bibfile, check = "warn", .Encoding = "UTF-8") %>%
as.data.frame()
# assign "categories" to the different types of publications
mypubs <- mypubs %>%
dplyr::mutate(
pubtype = dplyr::case_when(document_type == "Article" ~ "2",
document_type == "Article in Press" ~ "2",
document_type == "InProceedings" ~ "1",
document_type == "Proceedings" ~ "1",
document_type == "Conference" ~ "1",
document_type == "Conference Paper" ~ "1",
document_type == "MastersThesis" ~ "3",
document_type == "PhdThesis" ~ "3",
document_type == "Manual" ~ "4",
document_type == "TechReport" ~ "4",
document_type == "Book" ~ "5",
document_type == "InCollection" ~ "6",
document_type == "InBook" ~ "6",
document_type == "Misc" ~ "0",
TRUE ~ "0"))
# create a function which populates the md template based on the info
# about a publication
create_md <- function(x) {
# define a date and create filename by appending date and start of title
if (!is.na(x[["year"]])) {
x[["date"]] <- paste0(x[["year"]], "-01-01")
} else {
x[["date"]] <- "2999-01-01"
}
filename <- paste(x[["date"]], x[["title"]] %>%
str_replace_all(fixed(" "), "_") %>%
str_remove_all(fixed(":")) %>%
str_sub(1, 20) %>%
paste0(".md"), sep = "_")
# start writing
if (!file.exists(file.path(outfold, filename)) | overwrite) {
fileConn <- file.path(outfold, filename)
write("+++", fileConn)
# Title and date
write(paste0("title = \"", x[["title"]], "\""), fileConn, append = T)
write(paste0("date = \"", anydate(x[["date"]]), "\""), fileConn, append = T)
# Authors. Comma separated list, e.g. `["Bob Smith", "David Jones"]`.
auth_hugo <- str_replace_all(x["author"], " and ", "\", \"")
auth_hugo <- stringi::stri_trans_general(auth_hugo, "latin-ascii")
write(paste0("authors = [\"", auth_hugo,"\"]"), fileConn, append = T)
# Publication type. Legend:
# 0 = Uncategorized, 1 = Conference paper, 2 = Journal article
# 3 = Manuscript, 4 = Report, 5 = Book, 6 = Book section
write(paste0("publication_types = [\"", x[["pubtype"]],"\"]"),
fileConn, append = T)
# Publication details: journal, volume, issue, page numbers and doi link
publication <- x[["journal"]]
if (!is.na(x[["volume"]])) publication <- paste0(publication,
", (", x[["volume"]], ")")
if (!is.na(x[["number"]])) publication <- paste0(publication,
", ", x[["number"]])
if (!is.na(x[["pages"]])) publication <- paste0(publication,
", _pp. ", x[["pages"]], "_")
if (!is.na(x[["doi"]])) publication <- paste0(publication,
", ", paste0("https://doi.org/",
x[["doi"]]))
write(paste0("publication = \"", publication,"\""), fileConn, append = T)
write(paste0("publication_short = \"", publication,"\""),fileConn, append = T)
# Abstract and optional shortened version.
if (abstract) {
write(paste0("abstract = \"", x[["abstract"]],"\""), fileConn, append = T)
} else {
write("abstract = \"\"", fileConn, append = T)
}
write(paste0("abstract_short = \"","\""), fileConn, append = T)
# other possible fields are kept empty. They can be customized later by
# editing the created md
write("image_preview = \"\"", fileConn, append = T)
write("selected = false", fileConn, append = T)
write("projects = []", fileConn, append = T)
write("tags = []", fileConn, append = T)
#links
write("url_pdf = \"\"", fileConn, append = T)
write("url_preprint = \"\"", fileConn, append = T)
write("url_code = \"\"", fileConn, append = T)
write("url_dataset = \"\"", fileConn, append = T)
write("url_project = \"\"", fileConn, append = T)
write("url_slides = \"\"", fileConn, append = T)
write("url_video = \"\"", fileConn, append = T)
write("url_poster = \"\"", fileConn, append = T)
write("url_source = \"\"", fileConn, append = T)
#other stuff
write("math = true", fileConn, append = T)
write("highlight = true", fileConn, append = T)
# Featured image
write("[header]", fileConn, append = T)
write("image = \"\"", fileConn, append = T)
write("caption = \"\"", fileConn, append = T)
write("+++", fileConn, append = T)
}
}
# apply the "create_md" function over the publications list to generate
# the different "md" files.
apply(mypubs, FUN = function(x) create_md(x), MARGIN = 1)
}
Nothing fancy, here: I just use the
RefManageR package to read the BibTeX file, and then cycle over publications to create files properly formatted for hugo-academic use.Running the script
All is left is to run the script:
my_bibfile <- "/path/to/mybibtex.bib"
out_fold <- "/path/to/myoutfolder"
bibtex_2academic(bibffile = my_bibfile,
outfold = out_fold,
abstract = FALSE)- The
outfoldargument allows specifying where the generatedmdfiles will be saved. Though in the end they will have to be moved to foldercontent/publicationyou may want to save them at first in a different folder to be able to check them before trying to deploy. - The
abstractargument specifies whether to include the abstract in themdor not.
Running the script will give you files like this one:
+++
title = "Estimating canopy water content of poplar plantation from MIVIS data"
date = "2006-01-01"
authors = ["R. Colombo", "L. Busetto", "A. Marchesi", "M. Meroni", "C. Giardino"]
publication_types = ["1"]
publication = "AIP Conference Proceedings, (852), _pp. 242-249_, https://doi.org/10.1063/1.2349350"
publication_short = ""
abstract_short = ""
image_preview = ""
selected = false
projects = []
tags = []
url_pdf = ""
url_preprint = ""
url_code = ""
url_dataset = ""
url_project = ""
url_slides = ""
url_video = ""
url_poster = ""
url_source = ""
math = false
highlight = true
[header]
image = ""
caption = ""
+++
, where I tweaked a bit the hugo-academic format to include bibliographic info such as volume, number, pages and doi link. The files can then be further customized to include, for example, links to pdfs, images, etcetera.
After moving all the
md files to content/publication, the publications section of your hugo-academicsite will be auto-populated, and should look more or less like this: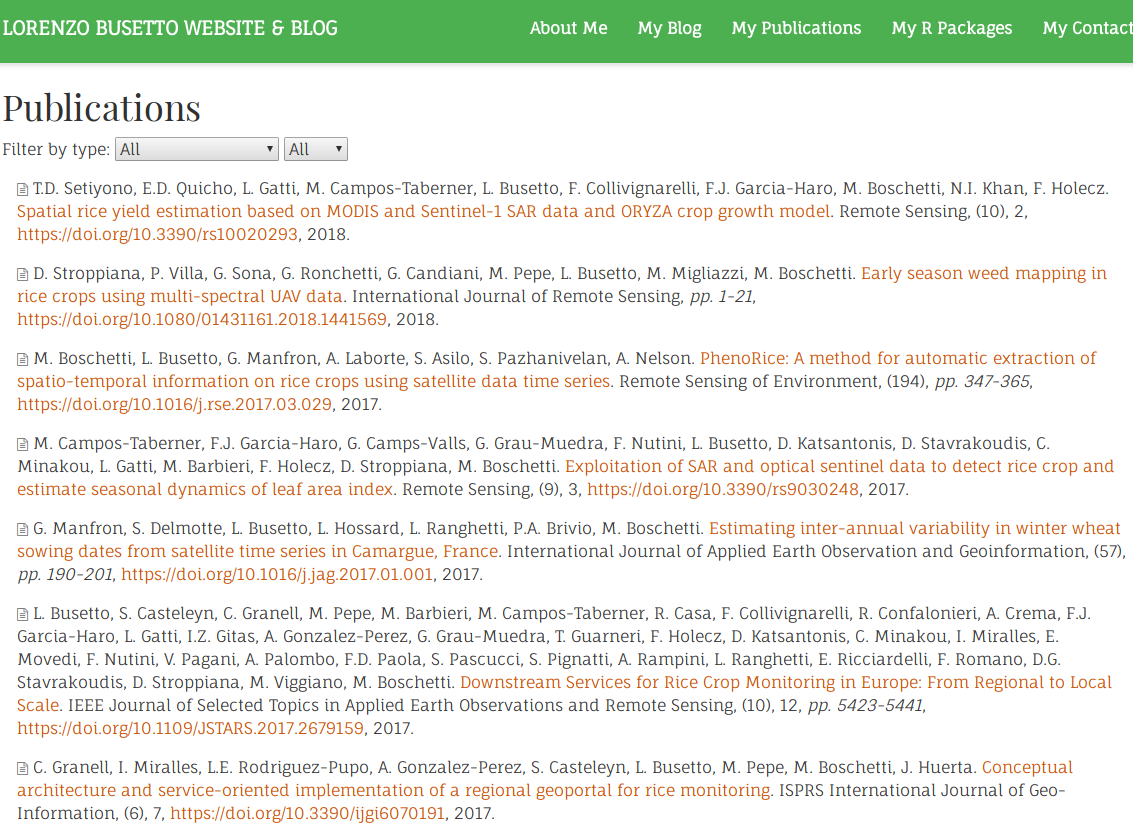
You can have a look at the final results on my (under construction) website here. I think it’s quite nice!
Final Notes
- My importing script is quite “quick and dirty”. It does not attempt to deal with special characters, and even substitutes accented letters with “bare” letters to avoid rendering problems. If someone more knowledgeable about encoding issues wants to try and improve it, I put it in this gist
- It may happen that your site will stop rendering when you put the new
mdfiles incontent/publication. If so, the reason is probably that you have some “strange” formatting in some of the files. Usual suspects would be unproperly recognized accents, math formulas or other special characters in the authors and abstract fields. You will have to look into each file and remove any offending areas. (It happened to me a lot before properly saving to UTF-8)
Thanks Lorenzo. This is great. I always wanted to code this problem but never had the free time. This should be useful for a lot of people. You should consider packaging it and send to CRAN.
ReplyDelete